PyLHC Submitter¶
See the API documentation for a detailed description of the code and the different parameters.
Note that the full functionality is only available under Linux with HTCondor configured, e.g. on CERN's lxplus service. Currently, due to lack of htcondor Python bindings on PypI for Windows and macOS, only local job execution is possible for these platforms.
Using the Job Submitter:¶
In the following examples, we will perform a tune sweep using a MAD-X mask. The simulations will be parametrized for both beams over a range of tunes in each plane. Parameters in the template script (see below) are indicated in the %(PARAMETER)s format.
The my_madx.mask Template File
! ----- Create Symlinks to Resources ----- !
option, warn, info;
system, "mkdir Outputdata";
system, "ln -fns /afs/cern.ch/eng/lhc/optics/V6.503 db5";
system, "ln -fns /afs/cern.ch/eng/lhc/optics/SLHCV1.0 slhc";
system, "ln -fns /afs/cern.ch/eng/lhc/optics/runII/2018 optics2018";
!option, -echo, warn, -info;
! ----- Make macros available ----- !
call, file="optics2018/toolkit/macro.madx";
! ----- Beam Options ----- !
mylhcbeam=%(BEAM)s; ! Will be replaced by values given to the submitter
qx=%(TUNEX)s; ! Will be replaced by values given to the submitter
qy=%(TUNEY)s; ! Will be replaced by values given to the submitter
emittance=3.75e-06;
n_part=1.0e10;
! ----- Set up Lattice ----- !
call, file="optics2018/lhc_as-built.seq"; ! LHC machine definition
! ----- Define the optics ----- !
call, file="optics2018/PROTON/opticsfile.22_ctpps2"; ! Optics to Round 30cm collision optics
! ----- Create Beams ----- !
beam, sequence=lhcb1, bv= 1, energy=NRJ, particle=proton, npart=n_part, kbunch=1, ex=emittance, ey=emittance;
beam, sequence=lhcb2, bv=-1, energy=NRJ, particle=proton, npart=n_part, kbunch=1, ex=emittance, ey=emittance;
! ----- Tune Matching ----- !
use, sequence=lhcb%(BEAM)s;
match, chrom;
global, q1=qx, q2=qy;
vary, name=dQx.b%(BEAM)s, step=1.0E-7; ! Will be replaced by values given to the submitter
vary, name=dQy.b%(BEAM)s, step=1.0E-7; ! Will be replaced by values given to the submitter
lmdif, calls=100, tolerance=1.0E-21;
endmatch;
! ----- Output Twiss ----- !
select, flag=twiss, clear;
select, flag=twiss, pattern="BPM", column=name,s,x,y,betx,bety,alfx,alfy,dx,dpx,mux,muy;
select, flag=twiss, pattern="M", column=name,s,x,y,betx,bety,alfx,alfy,dx,dpx,mux,muy;
select, flag=twiss, pattern="IP", column=name,s,x,y,betx,bety,alfx,alfy,dx,dpx,mux,muy;
twiss, chrom, file="Outputdata/b%(BEAM)s.twiss.tfs"; ! Will be replaced by values given to the submitter
! ----- Cleanup Symlinks ----- !
system, "unlink db5";
system, "unlink slhc";
system, "unlink optics2018";
Starting Studies from Python¶
Recommended Method
Starting studies from Python is the recommended method, especially with a high number of parameters. As we will see later on, other methods methods require all parameter values to be written down while the pythonic way allows for an easier and clearer definition of the parameter space.
The parametrizing of simulations and submission to HTCondor through Python is as simple as calling the main function of the submitter with the desired parameters. See below:
import numpy as np
from pylhc_submitter.job_submitter import main as htcondor_submit
if __name__ == "__main__":
htcondor_submit(
executable="madx", # default pointing to the latest MAD-X on afs
mask="my_madx.mask", # template to fill and execute MAD-X on
replace_dict=dict( # parameters to look for and replace in the template file
BEAM=[1, 2],
TUNEX=np.linspace(62.3, 62.32, 11).tolist(),
TUNEY=np.linspace(60.31, 60.33, 11).tolist(),
),
jobid_mask="b%(BEAM)d.qx%(TUNEX)s.qy%(TUNEY)s", # naming of the submitted jobfiles
working_directory="/afs/cern.ch/work/u/username/study.tune_sweep", # outputs
jobflavour="workday", # htcondor flavour
)
Starting Studies from a Config File¶
The same can also be achieved by specifying the previously called parameters in a config.ini file. It is worth noting that with any submission, the job_submitter will create such a file. See below the equivalent config.ini of the above Python example:
[DEFAULT]
executable="madx"
mask="my_madx.mask"
working_directory="/afs/cern.ch/work/u/username/study.tune_sweep"
replace_dict={
"BEAM": [1, 2],
"TUNEX": [62.3, 62.302, 62.304, 62.306, 62.308, 62.31, 62.312, 62.314, 62.316, 62.318, 62.32],
"TUNEY": [60.31, 60.312, 60.314, 60.316, 60.318, 60.32, 60.322, 60.324, 60.326, 60.328, 60.33],
}
jobid_mask="b%(BEAM)d.qx%(TUNEX)s.qy%(TUNEY)s"
jobflavour="workday"
The jobs are then started by calling the submitter on this file from the command line:
python -m pylhc_submitter.job_submitter --entry_cfg config.ini
Starting Studies from the Command Line¶
Users Beware
While doing so is possible, using a simple command line call is discouraged. As we will see in the example below, this method is not as clear as the previous ones, and prone to errors.
It is possible to skip the creation of a Python or a config.ini file completely when submitting, by providing parameters through the replace_dict flag at the command line. The above examples would be done through a (very lengthy) command line call as below:
python -m pylhc_submitter.job_submitter --executable madx --mask my_madx.mask --working_directory /afs/cern.ch/work/u/username/study.tune_sweep --replace_dict "{'BEAM': [1, 2], 'TUNEX': [62.3, 62.302, 62.304, 62.306, 62.308, 62.31, 62.312, 62.314, 62.316, 62.318, 62.32], 'TUNEY': [60.31, 60.312, 60.314, 60.316, 60.318, 60.32, 60.322, 60.324, 60.326, 60.328, 60.33]}" --jobid_mask b%(BEAM)d.qx%(TUNEX)s.qy%(TUNEY)s --jobflavour workday
Starting Studies with Mask Strings¶
Instead of using a mask file, job_submitter can also use a string as input for the executable. This can prove useful if the executable has a number of variable input parameters, i.e. executable --param1 X --param2 Y.
A simple mask string call using the config.ini input to the pylhc-submitter would look like this:
[DEFAULT]
executable="expr"
mask="%(SUMMAND1)s + %(SUMMAND2)s > Outputdata/result.txt"
working_directory="/afs/cern.ch/work/u/username/study.addition"
replace_dict={"SUMMAND1": [1, 2, 3, 4], "SUMMAND2": [6, 7, 8, 9]}
run_local=True
num_processes=4
Note that again, the user has to take care that the required results are saved in the correct job_output_dir. The mask string can be a more complicated multiline string, executing multiple commands.
Example Multiline Config File
[DEFAULT]
executable=None
mask="madx < Job%(SEED)s.madx\n
python -m analysis.main --method %(METHOD)s"
working_directory="/afs/cern.ch/work/u/username/study.multiline"
replace_dict={"SEED"= [1, 2, 3, 4], "METHOD"= ['fast', 'slow', 'new']}
run_local=True
num_processes=4
What Happens After Submitting¶
When submitting, the job_submitter determines jobs to be submitted to HTCondor through the inner product of the replace_dict: in our example each possible combination of the 3 given parameters. The script will issue several logging statements to keep the user updated, but here is what happens behind the scenes.
A folder structure is created in the given working directory, in which one will find:
- A Job.tfs file, a global summary dataframe in which each row corresponds to a set of parameters filled in for one submitted job:
- The first column,
JobId, is either numbering of the jobs or - in our case - the filled injobid_mask, - The next columns are the given parameters, here:
BEAM,TUNEXandTUNEY. They contain the value filled in the respective job-instance of said row, - The
JobDirectory,JobFileandShellScriptcolumns respectively contain the path to the job directory, the name of the filled mask file and the name of the script which are used to run the job.
- The first column,
- A queuehtc.sub file, the
HTCondorsubmission file used, which tellsHTCondorthe paths to all theShellScriptsto run. - Many sub-directories named according to the
jobid_mask, one per job, each with:- A
ShellScript.sh , which contains commands to create thejob_output_dirand themadxcommand to run the script, - A
JobFilewhich is the originalmask, parameter values filled in.
- A
Dry Runs
In case one runs the submitter with the dryrun flag, the execution stops here and prepared files are accessible, but nothing is sent to the HTCondor scheduler. One can use this option to perform a thorough check before using condor_submit on the created submission file queuehtc.sub.
Number of Jobs
As job_submitter creates a job for each combination in the inner product of the replace_dict parameters, the number of jobs can easily get high. In our example case, we are submitting beam * tunex * tuney = 2 * 11 * 11 = 242 jobs. It is worth noting that currently the maximum number of jobs to be submitted to HTCondor is set to 100k.
After submitting our tune sweep studies, we can check the status of our jobs via the condor_q (unless running locally). The output should look something like this:
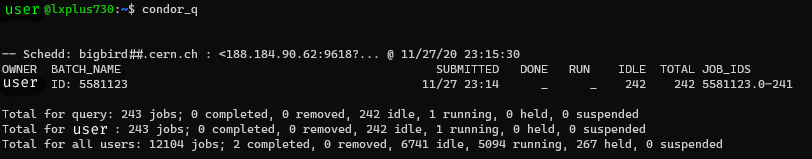
condor_q: Shows the jobs in the condor queue.When all jobs are done running, the htcondor.<jobid>.err, htcondor.<jobid>.log and htcondor.<jobid>.out files for the respective job are transferred back to the appropriate directories. Finally, for each job the job_output_dir, here containing the .twiss.tfs file, will be transferred back as well, ready for post-processing.
Checking for and Resubmitting Failed Jobs¶
To see if and which Jobs have failed, the same command as above can be rerun using resume_jobs and dryrun flags (or use them as parameters set to True). By default, Jobs are classified as successful if the specified job_output_dir is present.
To resubmit the failed jobs to HTCondor, simply rerun the call and omit the dryrun flag or set its parameter to False.
Refining the Resubmission
To further refine the resubmission, the script can also check for the presence of files in the job_output_dir by using the check_files flag. In the case of our tune sweep example, this would look like check_files=["*.twiss.tfs"].
Pitfalls and Hints¶
The ssh Option
When using the ssh option, the working directory needs to be accessible from both the local machine as well as the ssh server. In this case, ssh is only used for the final command submitting the jobs to HTCondor.
Running Locally
The run_local flag allows parallelizing jobs on your local machine through multiprocessing. This is only recommended for short jobs and for a small number of jobs, as the stress on your machine can be intense. The num_processes option allows to specify how many concurrent processes will run, by default 4.